- Record: found
- Abstract: found
- Article: found
hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing
Read this article at
Summary
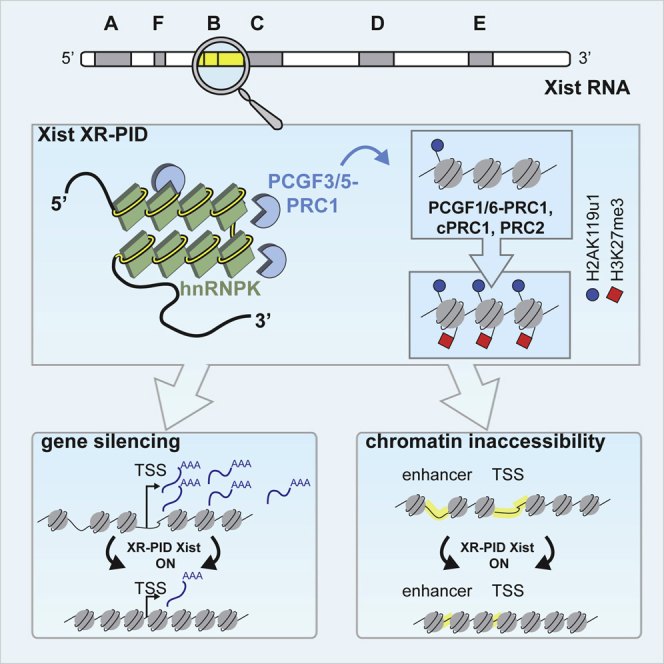
The Polycomb-repressive complexes PRC1 and PRC2 play a key role in chromosome silencing induced by the non-coding RNA Xist. Polycomb recruitment is initiated by the PCGF3/5-PRC1 complex, which catalyzes chromosome-wide H2A lysine 119 ubiquitylation, signaling recruitment of other PRC1 complexes, and PRC2. However, the molecular mechanism for PCGF3/5-PRC1 recruitment by Xist RNA is not understood. Here we define the Xist RNA Polycomb Interaction Domain (XR-PID), a 600 nt sequence encompassing the Xist B-repeat element. Deletion of XR-PID abolishes Xist-dependent Polycomb recruitment, in turn abrogating Xist-mediated gene silencing and reversing Xist-induced chromatin inaccessibility. We identify the RNA-binding protein hnRNPK as the principal XR-PID binding factor required to recruit PCGF3/5-PRC1. Accordingly, synthetically tethering hnRNPK to Xist RNA lacking XR-PID is sufficient for Xist-dependent Polycomb recruitment. Our findings define a key pathway for Polycomb recruitment by Xist RNA, providing important insights into mechanisms of chromatin modification by non-coding RNA.
Graphical Abstract
Highlights
Abstract
This study advances our understanding of the molecular mechanism of X chromosome inactivation in mammals, defining XR-PID, the critical element in Xist RNA that recruits Polycomb complexes to the inactive X chromosome, and further demonstrating that the RNA binding protein hnRNPK bridges XR-PID with the initiating Polycomb complex, PCGF3/5-PRC1.
Related collections
Most cited references33
- Record: found
- Abstract: not found
- Article: not found
Gene action in the X-chromosome of the mouse (Mus musculus L.).
- Record: found
- Abstract: found
- Article: not found
Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation.
- Record: found
- Abstract: found
- Article: not found