- Record: found
- Abstract: found
- Article: found
LSD1 Inhibition Promotes Epithelial Differentiation through Derepression of Fate-Determining Transcription Factors
Read this article at
SUMMARY
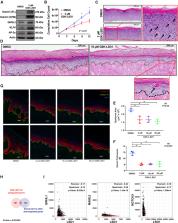
Self-renewing somatic tissues depend upon the proper balance of chromatin-modifying enzymes to coordinate progenitor cell maintenance and differentiation, disruption of which can promote carcinogenesis. As a result, drugs targeting the epigenome hold significant therapeutic potential. The histone demethylase, LSD1 (KDM1A), is overexpressed in numerous cancers, including epithelial cancers; however, its role in the skin is virtually unknown. Here we show that LSD1 directly represses master epithelial transcription factors that promote differentiation. LSD1 inhibitors block both LSD1 binding to chromatin and its catalytic activity, driving significant increases in H3K4 methylation and gene transcription of these fate-determining transcription factors. This leads to both premature epidermal differentiation and the repression of squamous cell carcinoma. Together these data highlight both LSD1’s role in maintaining the epidermal progenitor state and the potential of LSD1 inhibitors for the treatment of keratinocyte cancers, which collectively outnumber all other cancers combined.
Graphical Abstract

In Brief
Egolf et al. demonstrate that inhibition of the epigenetic regulator and histone demethylase, LSD1, promotes activation of the epidermal differentiation transcriptional program and, in turn, represses the invasion of cutaneous squamous cell carcinoma, one of the most common of all human cancers.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
NIH Image to ImageJ: 25 years of image analysis.
- Record: found
- Abstract: found
- Article: not found
BEDTools: The Swiss-Army Tool for Genome Feature Analysis.
- Record: found
- Abstract: found
- Article: not found