- Record: found
- Abstract: found
- Article: found
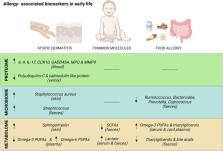
Allergy-associated biomarkers in early life identified by Omics techniques
Read this article at
Abstract
The prevalence and severity of allergic diseases have increased over the last 30 years. Understanding the mechanisms responsible for these diseases is a major challenge in current allergology, as it is crucial for the transition towards precision medicine, which encompasses predictive, preventive, and personalized strategies. The urge to identify predictive biomarkers of allergy at early stages of life is crucial, especially in the context of major allergic diseases such as food allergy and atopic dermatitis. Identifying these biomarkers could enhance our understanding of the immature immune responses, improve allergy handling at early ages and pave the way for preventive and therapeutic approaches. This minireview aims to explore the relevance of three biomarker categories (proteome, microbiome, and metabolome) in early life. First, levels of some proteins emerge as potential indicators of mucosal health and metabolic status in certain allergic diseases. Second, bacterial taxonomy provides insight into the composition of the microbiota through high-throughput sequencing methods. Finally, metabolites, representing the end products of bacterial and host metabolic activity, serve as early indicators of changes in microbiota and host metabolism. This information could help to develop an extensive identification of biomarkers in AD and FA and their potential in translational personalized medicine in early life.
Related collections
Most cited references172
- Record: found
- Abstract: found
- Article: not found
The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis.
- Record: found
- Abstract: found
- Article: not found
Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns.
- Record: found
- Abstract: found
- Article: not found
