- Record: found
- Abstract: found
- Article: found
Heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs) function as endocytic receptors for an internalizing anti-nucleic acid antibody
Read this article at
Abstract
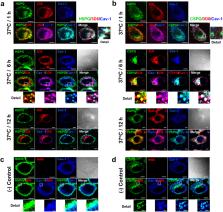
A subset of monoclonal anti-DNA autoantibodies enters a variety of living cells. Here, we aimed to identify the endocytic receptors recognized by an internalizing anti-nucleic acid autoantibody, the 3D8 single-chain variable fragment (scFv). We found that cell surface binding and internalization of 3D8 scFv were inhibited markedly in soluble heparan sulfate (HS)/chondroitin sulfate (CS)-deficient or -removed cells and in the presence of soluble HS and CS. 3D8 scFv colocalized intracellularly with either HS proteoglycans (HSPGs) or CSPGs in HeLa cells. 3D8 scFv was co-endocytosed and co-precipitated with representative individual HSPG and CSPG molecules: syndecan-2 (a transmembrane HSPG), glypican-3 (a glycosylphosphatidylinositol (GPI)-anchored HSPG); CD44 (a transmembrane CSPG); and brevican (a GPI-anchored CSPG). Collected data indicate that 3D8 scFv binds to the negatively charged sugar chains of both HSPGs and CSPGs and is then internalized along with these molecules, irrespective of how these proteoglycans are associated with the cell membrane. This is the first study to show that anti-DNA antibodies enter cells via both HSPGs and CSPGs simultaneously. The data may aid understanding of endocytic receptors that bind anti-DNA autoantibodies. The study also provides insight into potential cell membrane targets for macromolecular delivery.
Related collections
Most cited references42
- Record: found
- Abstract: found
- Article: not found
Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity.
- Record: found
- Abstract: found
- Article: not found
Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds.
- Record: found
- Abstract: found
- Article: not found
