- Record: found
- Abstract: found
- Article: found
Burkholderiaceae Are Key Acetate Assimilators During Complete Denitrification in Acidic Cryoturbated Peat Circles of the Arctic Tundra
Read this article at
Abstract
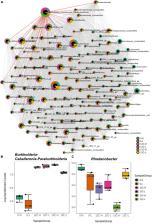
Cryoturbated peat circles (pH 4) in the Eastern European Tundra harbor up to 2 mM pore water nitrate and emit the greenhouse gas N 2O like heavily fertilized agricultural soils in temperate regions. The main process yielding N 2O under oxygen limited conditions is denitrification, which is the sequential reduction of nitrate/nitrite to N 2O and/or N 2. N 2O reduction to N 2 is impaired by pH < 6 in classical model denitrifiers and many environments. Key microbes of peat circles are important but largely unknown catalysts for C- and N-cycling associated N 2O fluxes. Thus, we hypothesized that the peat circle community includes hitherto unknown taxa and is essentially unable to efficiently perform complete denitrification, i.e., reduce N 2O, due to a low in situ pH. 16S rRNA analysis indicated a diverse active community primarily composed of the bacterial class-level taxa Alphaproteobacteria, Acidimicrobiia, Acidobacteria, Verrucomicrobiae, and Bacteroidia, as well as archaeal Nitrososphaeria. Euryarchaeota were not detected. 13C 2- and 12C 2-acetate supplemented anoxic microcosms with endogenous nitrate and acetylene at an in situ near pH of 4 were used to assess acetate dependent carbon flow, denitrification and N 2O production. Initial nitrate and acetate were consumed within 6 and 11 days, respectively, and primarily converted to CO 2 and N 2, suggesting complete acetate fueled denitrification at acidic pH. Stable isotope probing coupled to 16S rRNA analysis via Illumina MiSeq amplicon sequencing identified acetate consuming key players of the family Burkholderiaceae during complete denitrification correlating with Rhodanobacter spp. The archaeal community consisted primarily of ammonia-oxidizing Archaea of Nitrososphaeraceae, and was stable during the incubation. The collective data indicate that peat circles (i) host acid-tolerant denitrifiers capable of complete denitrification at pH 4–5.5, (ii) other parameters like carbon availability rather than pH are possible reasons for high N 2O emissions in situ, and (iii) Burkholderiaceae are responsive key acetate assimilators co-occurring with Rhodanobacter sp. during denitrification, suggesting both organisms being associated with acid-tolerant denitrification in peat circles.
Related collections
Most cited references103

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2

- Record: found
- Abstract: found
- Article: found
The SILVA ribosomal RNA gene database project: improved data processing and web-based tools
- Record: found
- Abstract: found
- Article: not found