- Record: found
- Abstract: found
- Article: found
Probiotic Bifidobacterium longum alters gut luminal metabolism through modification of the gut microbial community
Read this article at
Abstract
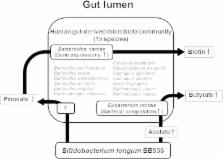
Probiotics are well known as health-promoting agents that modulate intestinal microbiota. However, the molecular mechanisms underlying this effect remain unclear. Using gnotobiotic mice harboring 15 strains of predominant human gut-derived microbiota (HGM), we investigated the effects of Bifidobacterium longum BB536 (BB536-HGM) supplementation on the gut luminal metabolism. Nuclear magnetic resonance (NMR)-based metabolomics showed significantly increased fecal levels of pimelate, a precursor of biotin, and butyrate in the BB536-HGM group. In addition, the bioassay revealed significantly elevated fecal levels of biotin in the BB536-HGM group. Metatranscriptomic analysis of fecal microbiota followed by an in vitro bioassay indicated that the elevated biotin level was due to an alteration in metabolism related to biotin synthesis by Bacteroides caccae in this mouse model. Furthermore, the proportion of Eubacterium rectale, a butyrate producer, was significantly higher in the BB536-HGM group than in the group without B. longum BB536 supplementation. Our findings help to elucidate the molecular basis underlying the effect of B. longum BB536 on the gut luminal metabolism through its interactions with the microbial community.
Related collections
Most cited references34
- Record: found
- Abstract: found
- Article: not found
How glycan metabolism shapes the human gut microbiota.
- Record: found
- Abstract: found
- Article: not found
Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla.

- Record: found
- Abstract: found
- Article: found
