- Record: found
- Abstract: found
- Article: found
The Neonatal and Adult Human Testis Defined at the Single-Cell Level
Read this article at
SUMMARY
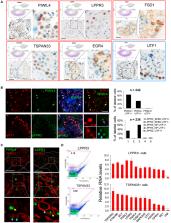
Spermatogenesis has been intensely studied in rodents but remains poorly understood in humans. Here, we used single-cell RNA sequencing to analyze human testes. Clustering analysis of neonatal testes reveals several cell subsets, including cell populations with characteristics of primordial germ cells (PGCs) and spermatogonial stem cells (SSCs). In adult testes, we identify four undifferentiated spermatogonia (SPG) clusters, each of which expresses specific marker genes. We identify protein markers for the most primitive SPG state, allowing us to purify this likely SSC-enriched cell subset. We map the timeline of male germ cell development from PGCs through fetal germ cells to differentiating adult SPG stages. We also define somatic cell subsets in both neonatal and adult testes and trace their developmental trajectories. Our data provide a blueprint of the developing human male germline and supporting somatic cells. The PGC-like and SSC markers are candidates to be used for SSC therapy to treat infertility.
Graphical Abstract

In Brief
Sohni et al. use scRNA-seq analysis to define cell subsets in the human testis. Highlights include the identification of primordial germ cell- and spermatogonial stem cell-like cell subsets in neonatal testes, numerous undifferentiated spermatogonial cell states in adult testes, and somatic cell subsets in both neonatal and adult testes.
Related collections
Most cited references48

- Record: found
- Abstract: found
- Article: found
The adult human testis transcriptional cell atlas
- Record: found
- Abstract: found
- Article: not found
The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids
- Record: found
- Abstract: not found
- Article: not found