- Record: found
- Abstract: found
- Article: found
Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation
Read this article at
Abstract
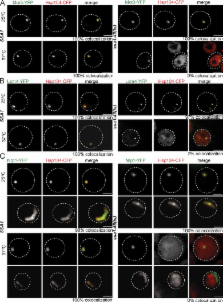
The Hsp70 system recruits ClpB/Hsp104 to the surface of stress-induced protein aggregates and prion fibrils.
Abstract
Hsp100 and Hsp70 chaperones in bacteria, yeast, and plants cooperate to reactivate aggregated proteins. Disaggregation relies on Hsp70 function and on ATP-dependent threading of aggregated polypeptides through the pore of the Hsp100 AAA + hexamer. In yeast, both chaperones also promote propagation of prions by fibril fragmentation, but their functional interplay is controversial. Here, we demonstrate that Hsp70 chaperones were essential for species-specific targeting of their Hsp100 partner chaperones ClpB and Hsp104, respectively, to heat-induced protein aggregates in vivo. Hsp70 inactivation in yeast also abrogated Hsp104 targeting to almost all prions tested and reduced fibril mobility, which indicates that fibril fragmentation by Hsp104 requires Hsp70. The Sup35 prion was unique in allowing Hsp70-independent association of Hsp104 via its N-terminal domain, which, however, was nonproductive. Hsp104 overproduction even outcompeted Hsp70 for Sup35 prion binding, which explains why this condition prevented Sup35 fragmentation and caused prion curing. Our findings indicate a conserved mechanism of Hsp70–Hsp100 cooperation at the surface of protein aggregates and prion fibrils.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins.
- Record: found
- Abstract: found
- Article: not found
Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104.
- Record: found
- Abstract: not found
- Article: not found