- Record: found
- Abstract: found
- Article: found
Pyramiding stacking of multigenes (PSM): a simple, flexible and efficient multigene stacking system based on Gibson assembly and gateway cloning
Read this article at
Abstract
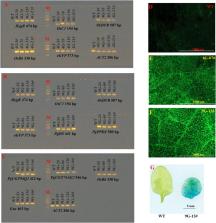
Genetic engineering of complex metabolic pathways and multiple traits often requires the introduction of multiple genes. The construction of plasmids carrying multiple DNA fragments plays a vital role in these processes. In this study, the Gibson assembly and Gateway cloning combined Pyramiding Stacking of Multigenes (PSM) system was developed to assemble multiple transgenes into a single T-DNA. Combining the advantages of Gibson assembly and Gateway cloning, the PSM system uses an inverted pyramid stacking route and allows fast, flexible and efficient stacking of multiple genes into a binary vector. The PSM system contains two modular designed entry vectors (each containing two different attL sites and two selectable markers) and one Gateway-compatible destination vector (containing four attR sites and two negative selection markers). The target genes are primarily assembled into the entry vectors via two parallel rounds of Gibson assembly reactions. Then, the cargos in the entry constructs are integrated into the destination vector via a single tube Gateway LR reaction. To demonstrate PSM’s capabilities, four and nine gene expression cassettes were respectively assembled into the destination vector to generate two binary expression vectors. The transgenic analysis of these constructs in Arabidopsis demonstrated the reliability of the constructs generated by PSM. Due to its flexibility, simplicity and versatility, PSM has great potential for genetic engineering, synthetic biology and the improvement of multiple traits.
Related collections
Most cited references42
- Record: found
- Abstract: not found
- Article: not found
Enzymatic assembly of DNA molecules up to several hundred kilobases.
- Record: found
- Abstract: found
- Article: not found
Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana
- Record: found
- Abstract: found
- Article: not found