- Record: found
- Abstract: found
- Article: found
Cell type phylogenetics informs the evolutionary origin of echinoderm larval skeletogenic cell identity
Read this article at
Abstract
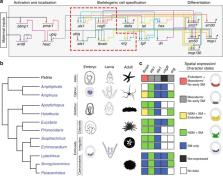
The multiplicity of cell types comprising multicellular organisms begs the question as to how cell type identities evolve over time. Cell type phylogenetics informs this question by comparing gene expression of homologous cell types in distantly related taxa. We employ this approach to inform the identity of larval skeletogenic cells of echinoderms, a clade for which there are phylogenetically diverse datasets of spatial gene expression patterns. We determined ancestral spatial expression patterns of alx1, ets1, tbr, erg, and vegfr, key components of the skeletogenic gene regulatory network driving identity of the larval skeletogenic cell. Here we show ancestral state reconstructions of spatial gene expression of extant eleutherozoan echinoderms support homology and common ancestry of echinoderm larval skeletogenic cells. We propose larval skeletogenic cells arose in the stem lineage of eleutherozoans during a cell type duplication event that heterochronically activated adult skeletogenic cells in a topographically distinct tissue in early development.
Abstract
Eric Erkenbrack and Jeffrey Thompson have performed ancestral state reconstructions of echinoderm larval skeletogenic cells to examine their relatedness and provide insights into cell-type evolution. They propose that larval skeletogenic cells originated in the stem lineage of eleutherozoans.
Related collections
Most cited references91
- Record: found
- Abstract: not found
- Article: not found
Detecting Correlated Evolution on Phylogenies: A General Method for the Comparative Analysis of Discrete Characters
- Record: found
- Abstract: found
- Article: not found
The selection and function of cell type-specific enhancers.
- Record: found
- Abstract: not found
- Article: not found