- Record: found
- Abstract: found
- Article: found
Inferring time-dependent population growth rates in cell cultures undergoing adaptation
Read this article at
Abstract
Background
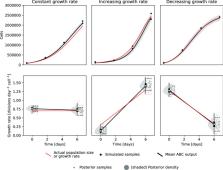
The population growth rate is an important characteristic of any cell culture. During sustained experiments, the growth rate may vary due to competition or adaptation. For instance, in presence of a toxin or a drug, an increasing growth rate indicates that the cells adapt and become resistant. Consequently, time-dependent growth rates are fundamental to follow on the adaptation of cells to a changing evolutionary landscape. However, as there are no tools to calculate the time-dependent growth rate directly by cell counting, it is common to use only end point measurements of growth rather than tracking the growth rate continuously.
Results
We present a computer program for inferring the growth rate over time in suspension cells using nothing but cell counts, which can be measured non-destructively. The program was tested on simulated and experimental data. Changes were observed in the initial and absolute growth rates, betraying resistance and adaptation.
Conclusions
For experiments where adaptation is expected to occur over a longer time, our method provides a means of tracking growth rates using data that is normally collected anyhow for monitoring purposes. The program and its documentation are freely available at https://github.com/Sandalmoth/ratrack under the permissive zlib license.
Related collections
Most cited references25
- Record: found
- Abstract: found
- Article: not found
Snakemake--a scalable bioinformatics workflow engine.
- Record: found
- Abstract: found
- Article: found
Raincloud plots: a multi-platform tool for robust data visualization
- Record: found
- Abstract: found
- Article: found