- Record: found
- Abstract: found
- Article: found
Hybrid Dysgenesis in Drosophila simulans Associated with a Rapid Invasion of the P-Element
Read this article at
Abstract
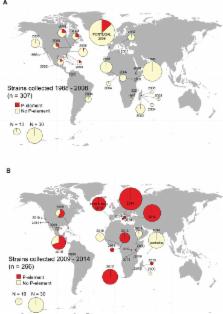
In a classic example of the invasion of a species by a selfish genetic element, the P-element was horizontally transferred from a distantly related species into Drosophila melanogaster. Despite causing ‘hybrid dysgenesis’, a syndrome of abnormal phenotypes that include sterility, the P-element spread globally in the course of a few decades in D. melanogaster. Until recently, its sister species, including D. simulans, remained P-element free. Here, we find a hybrid dysgenesis-like phenotype in the offspring of crosses between D. simulans strains collected in different years; a survey of 181 strains shows that around 20% of strains induce hybrid dysgenesis. Using genomic and transcriptomic data, we show that this dysgenesis-inducing phenotype is associated with the invasion of the P-element. To characterize this invasion temporally and geographically, we survey 631 D. simulans strains collected on three continents and over 27 years for the presence of the P-element. We find that the D. simulans P-element invasion occurred rapidly and nearly simultaneously in the regions surveyed, with strains containing P-elements being rare in 2006 and common by 2014. Importantly, as evidenced by their resistance to the hybrid dysgenesis phenotype, strains collected from the latter phase of this invasion have adapted to suppress the worst effects of the P-element.
Author Summary
Some genes perform necessary organismal functions, others hijack the cellular machinery to replicate themselves, potentially harming the host in the process. These ‘selfish genes’ can spread through genomes and species; as a result, eukaryotic genomes are typically saddled with large amounts of parasitic DNA. Here, we chronicle the surprisingly rapid global spread of a selfish transposable element through a close relative of the genetic model, Drosophila melanogaster. We see that, as it spreads, the transposable element is associated with damaging effects, including sterility, but that the flies quickly adapt to the negative consequences of the transposable element.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
Selfish genes, the phenotype paradigm and genome evolution.
- Record: found
- Abstract: found
- Article: not found
Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary.
- Record: found
- Abstract: found
- Article: not found