- Record: found
- Abstract: found
- Article: not found
Atomic Resolution Insights into Curli Fiber Biogenesis
Read this article at
Summary
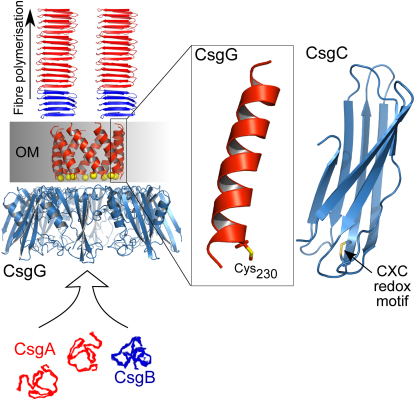
Bacteria produce functional amyloid fibers called curli in a controlled, noncytotoxic manner. These extracellular fimbriae enable biofilm formation and promote pathogenicity. Understanding curli biogenesis is important for appreciating microbial lifestyles and will offer clues as to how disease-associated human amyloid formation might be ameliorated. Proteins encoded by the curli specific genes ( csgA-G) are required for curli production. We have determined the structure of CsgC and derived the first structural model of the outer-membrane subunit translocator CsgG. Unexpectedly, CsgC is related to the N-terminal domain of DsbD, both in structure and oxido-reductase capability. Furthermore, we show that CsgG belongs to the nascent class of helical outer-membrane macromolecular exporters. A cysteine in a CsgG transmembrane helix is a potential target of CsgC, and mutation of this residue influences curli assembly. Our study provides the first high-resolution structural insights into curli biogenesis.
Abstract
Highlights
► CsgC is related to the redox-active N-terminal domain of DsbD ► Outer-membrane transporter CsgG inserts into the membrane via an α-helical oligomer ► The transmembrane domain of CsgG is vital for pore forming and curli assembly ► CsgC appears to affect CsgG pore behavior and biofilm formation
Related collections
Most cited references47
- Record: found
- Abstract: found
- Article: not found
The PSIPRED protein structure prediction server.
- Record: found
- Abstract: found
- Article: not found
REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use.
- Record: found
- Abstract: found
- Article: not found