- Record: found
- Abstract: found
- Article: found
Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning
Read this article at
Abstract
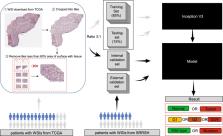
Hepatocellular carcinoma (HCC) is the most common subtype of liver cancer, and assessing its histopathological grade requires visual inspection by an experienced pathologist. In this study, the histopathological H&E images from the Genomic Data Commons Databases were used to train a neural network (inception V3) for automatic classification. According to the evaluation of our model by the Matthews correlation coefficient, the performance level was close to the ability of a 5-year experience pathologist, with 96.0% accuracy for benign and malignant classification, and 89.6% accuracy for well, moderate, and poor tumor differentiation. Furthermore, the model was trained to predict the ten most common and prognostic mutated genes in HCC. We found that four of them, including CTNNB1, FMN2, TP53, and ZFX4, could be predicted from histopathology images, with external AUCs from 0.71 to 0.89. The findings demonstrated that convolutional neural networks could be used to assist pathologists in the classification and detection of gene mutation in liver cancer.
Related collections
Most cited references21
- Record: found
- Abstract: found
- Article: not found
Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning
- Record: found
- Abstract: found
- Article: not found
Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer
- Record: found
- Abstract: found
- Article: found