- Record: found
- Abstract: found
- Article: not found
Evolutionary and molecular foundations of multiple contemporary functions of the nitroreductase superfamily
Read this article at
Significance
Functionally diverse enzyme superfamilies are sets of homologs that conserve a structural fold and mechanistic details but perform various distinct chemical reactions. What are the evolutionary routes by which ancestral proteins diverge to produce extant enzymes? We present an approach that combines experimental data with computational tools to trace these sequence–structure–function transitions in a model system, the functionally diverse flavin mononucleotide-dependent nitroreductases (NTRs). Our results suggest an evolutionary model in which contemporary NTR classes have diverged in a radial manner from a minimal flavin-binding scaffold via insertions at key positions and fixation of functional residues, yielding the reaction versatility of contemporary enzymes. These principles will facilitate rational design of NTRs and advance general approaches for delineating the emergence of functional diversity in enzyme superfamilies.
Abstract
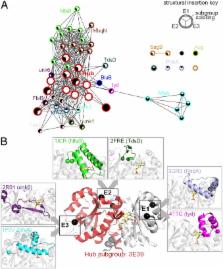
Insight regarding how diverse enzymatic functions and reactions have evolved from ancestral scaffolds is fundamental to understanding chemical and evolutionary biology, and for the exploitation of enzymes for biotechnology. We undertook an extensive computational analysis using a unique and comprehensive combination of tools that include large-scale phylogenetic reconstruction to determine the sequence, structural, and functional relationships of the functionally diverse flavin mononucleotide-dependent nitroreductase (NTR) superfamily (>24,000 sequences from all domains of life, 54 structures, and >10 enzymatic functions). Our results suggest an evolutionary model in which contemporary subgroups of the superfamily have diverged in a radial manner from a minimal flavin-binding scaffold. We identified the structural design principle for this divergence: Insertions at key positions in the minimal scaffold that, combined with the fixation of key residues, have led to functional specialization. These results will aid future efforts to delineate the emergence of functional diversity in enzyme superfamilies, provide clues for functional inference for superfamily members of unknown function, and facilitate rational redesign of the NTR scaffold.
Related collections
Most cited references82
- Record: found
- Abstract: not found
- Article: not found
Basic Local Alignment Search Tool
- Record: found
- Abstract: found
- Article: found
Maps of random walks on complex networks reveal community structure
- Record: found
- Abstract: found
- Article: not found
