- Record: found
- Abstract: found
- Article: found
Comparison of paired cerebrospinal fluid and serum cell‐free mitochondrial and nuclear DNA with copy number and fragment length
Read this article at
Abstract
Background
Most studies on cell‐free DNA (cfDNA) were only for single body fluids; however, the differences in cfDNA distribution between two body fluids are rarely reported. Hence, in this work, we compared the differences in cfDNA distribution between cerebrospinal fluid (CSF) and serum of patients with brain‐related diseases.
Methods
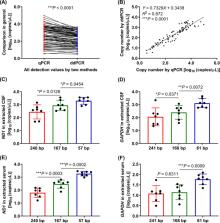
The fragment length of cfDNA was determined by using Agilent 2100 Bioanalyzer. The copy numbers of cell‐free mitochondrial DNA (cf‐mtDNA) and cell‐free nuclear DNA (cf‐nDNA) were determined by using real‐time quantitative PCR (qPCR) and droplet digital PCR (ddPCR) with three pairs of mitochondrial ND1 and nuclear GAPDH primers, respectively.
Results
There were short (~60 bp), medium (~167 bp), and long (>250 bp) cfDNA fragment length distributions totally obtained from CSF and serum using Agilent 2100 Bioanalyzer. The results of both qPCR and ddPCR confirmed the existence of these three cfDNA fragment ranges in CSF and serum. According to qPCR, the copy numbers of long cf‐mtDNA, medium, and long cf‐nDNA in CSF were significantly higher than in paired serum. In CSF, only long cf‐mtDNA's copy numbers were higher than long cf‐nDNA. But in serum, the copy numbers of medium and long cf‐mtDNA were higher than the corresponding cf‐nDNA.
Related collections
Most cited references36

- Record: found
- Abstract: found
- Article: found
Origins, structures, and functions of circulating DNA in oncology
- Record: found
- Abstract: found
- Article: not found
Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation.
- Record: found
- Abstract: found
- Article: not found