- Record: found
- Abstract: found
- Article: not found
Microfluidic Control of Cell Pairing and Fusion
Abstract
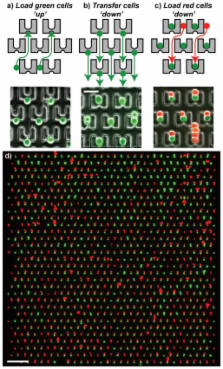
Cell fusion has been used for many different purposes, including generation of hybridomas and reprogramming of somatic cells. The fusion step represents the key event in initiation of these procedures. Standard fusion techniques, however, provide poor and random cell contact, leading to low yields. We present here a microfluidic device to trap and properly pair thousands of cells. Using this device we were able to pair different cell types, including fibroblasts, mouse embryonic stem cells (mESCs), and myeloma cells, achieving pairing efficiencies up to 70%. The device is compatible with both chemical and electrical fusion protocols. We observed that electrical fusion was more efficient than chemical fusion, with membrane reorganization efficiencies of up to 89%. We achieved greater than 50% properly paired and fused cells over the entire device, 5× greater than a commercial electrofusion chamber, and were able to observe reprogramming in hybrids between mESCs and mouse embryonic fibroblasts.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Viable offspring derived from fetal and adult mammalian cells.
- Record: found
- Abstract: found
- Article: not found
In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state.
- Record: found
- Abstract: found
- Article: not found

