- Record: found
- Abstract: found
- Article: found
Glyco-engineered cell line and computational docking studies reveals enterotoxigenic Escherichia coli CFA/I fimbriae bind to Lewis a glycans
Read this article at
Abstract
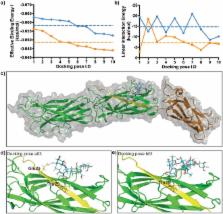
We have previously reported clinical data to suggest that colonization factor I (CFA/I) fimbriae of enterotoxigenic Escherichia coli (ETEC) can bind to Lewis a (Le a), a glycan epitope ubiquitous in the small intestinal mucosa of young children (<2 years of age), and individuals with a genetic mutation of FUT2. To further elucidate the physiological binding properties of this interaction, we engineered Chinese Hamster Ovary (CHO-K1) cells to express Le a or Le b determinants on both N- and O-glycans. We used our glyco-engineered CHO-K1 cell lines to demonstrate that CfaB, the major subunit of ETEC CFA/I fimbriae, as well as four related ETEC fimbriae, bind more to our CHO-K1 cell-line expressing Le a, compared to cells carrying Le b or the CHO-K1 wild-type glycan phenotype. Furthermore, using in-silico docking analysis, we predict up to three amino acids (Glu 25, Asn 27, Thr 29) found in the immunoglobulin (Ig)-like groove region of CfaB of CFA/I and related fimbriae, could be important for the preferential and higher affinity binding of CFA/I fimbriae to the potentially structurally flexible Le a glycan. These findings may lead to a better molecular understanding of ETEC pathogenesis, aiding in the development of vaccines and/or anti-infection therapeutics.
Related collections
Most cited references27
- Record: found
- Abstract: found
- Article: not found
A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression.
- Record: found
- Abstract: found
- Article: not found
Status of vaccine research and development for enterotoxigenic Escherichia coli.
- Record: found
- Abstract: found
- Article: not found