- Record: found
- Abstract: found
- Article: found
Reversing the miRNA -5p/-3p stoichiometry reveals physiological roles and targets of miR-140 miRNAs
Read this article at
Abstract
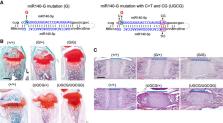
The chondrocyte-specific miR-140 miRNAs are necessary for normal endochondral bone growth in mice. miR-140 deficiency causes dwarfism and craniofacial deformity. However, the physiologically important targets of miR-140 miRNAs are still unclear. The miR-140 gene ( Mir140) encodes three chondrocyte-specific microRNAs, miR-140-5p, derived from the 5′ strand of primary miR-140, and miR140-3p.1 and -3p.2, derived from the 3′ strand of primary miR-140. miR-140-3p miRNAs are 10 times more abundant than miR-140-5p likely due to the nonpreferential loading of miR-140-5p to Argonaute proteins. To differentiate the role of miR-140-5p and -3p miRNAs in endochondral bone development, two distinct mouse models, miR140-C > T, in which the first nucleotide of miR-140-5p was altered from cytosine to uridine, and miR140-CG, where the first two nucleotides of miR-140-3p were changed to cytosine and guanine, were created. These changes are expected to alter Argonaute protein loading preference of -5p and -3p to increase -5p loading and decrease -3p loading without changing the function of miR140-5p. These models presented a mild delay in epiphyseal development with delayed chondrocyte maturation. Using RNA-sequencing analysis of the two models, direct targets of miR140-5p, including Wnt11, were identified. Disruption of the predicted miR140-5p binding site in the 3′ untranslated region of Wnt11 was shown to increase Wnt11 mRNA expression and caused a modest acceleration of epiphyseal development. These results show that the relative abundance of miRNA-5p and -3p can be altered by changing the first nucleotide of miRNAs in vivo, and this method can be useful to identify physiologically important miRNA targets.
Related collections
Most cited references29

- Record: found
- Abstract: found
- Article: found
Predicting effective microRNA target sites in mammalian mRNAs
- Record: found
- Abstract: found
- Article: not found