- Record: found
- Abstract: found
- Article: found
Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth
Read this article at
Abstract
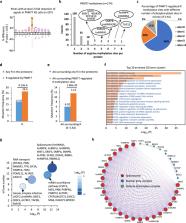
Numerous substrates have been identified for Type I and II arginine methyltransferases (PRMTs). However, the full substrate spectrum of the only type III PRMT, PRMT7, and its connection to type I and II PRMT substrates remains unknown. Here, we use mass spectrometry to reveal features of PRMT7-regulated methylation. We find that PRMT7 predominantly methylates a glycine and arginine motif; multiple PRMT7-regulated arginine methylation sites are close to phosphorylations sites; methylation sites and proximal sequences are vulnerable to cancer mutations; and methylation is enriched in proteins associated with spliceosome and RNA-related pathways. We show that PRMT4/5/7-mediated arginine methylation regulates hnRNPA1 binding to RNA and several alternative splicing events. In breast, colorectal and prostate cancer cells, PRMT4/5/7 are upregulated and associated with high levels of hnRNPA1 arginine methylation and aberrant alternative splicing. Pharmacological inhibition of PRMT4/5/7 suppresses cancer cell growth and their co-inhibition shows synergistic effects, suggesting them as targets for cancer therapy.
Abstract
Arginine methyltransferases (PRMTs) are involved in the regulation of various physiological and pathological conditions. Using proteomics, the authors here profile the methylation substrates of PRMTs 4, 5 and 7 and characterize the roles of these enzymes in cancer-associated splicing regulation.
Related collections
Most cited references80

- Record: found
- Abstract: found
- Article: found
Metascape provides a biologist-oriented resource for the analysis of systems-level datasets
- Record: found
- Abstract: found
- Article: not found
Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks.
- Record: found
- Abstract: found
- Article: found