- Record: found
- Abstract: found
- Article: found
A Massively Parallel Sequencing Approach Uncovers Ancient Origins and High Genetic Variability of Endangered Przewalski's Horses
Read this article at
Abstract
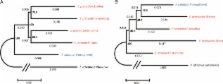
The endangered Przewalski's horse is the closest relative of the domestic horse and is the only true wild horse species surviving today. The question of whether Przewalski's horse is the direct progenitor of domestic horse has been hotly debated. Studies of DNA diversity within Przewalski's horses have been sparse but are urgently needed to ensure their successful reintroduction to the wild. In an attempt to resolve the controversy surrounding the phylogenetic position and genetic diversity of Przewalski's horses, we used massively parallel sequencing technology to decipher the complete mitochondrial and partial nuclear genomes for all four surviving maternal lineages of Przewalski's horses. Unlike single-nucleotide polymorphism (SNP) typing usually affected by ascertainment bias, the present method is expected to be largely unbiased. Three mitochondrial haplotypes were discovered—two similar ones, haplotypes I/II, and one substantially divergent from the other two, haplotype III. Haplotypes I/II versus III did not cluster together on a phylogenetic tree, rejecting the monophyly of Przewalski's horse maternal lineages, and were estimated to split 0.117–0.186 Ma, significantly preceding horse domestication. In the phylogeny based on autosomal sequences, Przewalski's horses formed a monophyletic clade, separate from the Thoroughbred domestic horse lineage. Our results suggest that Przewalski's horses have ancient origins and are not the direct progenitors of domestic horses. The analysis of the vast amount of sequence data presented here suggests that Przewalski's and domestic horse lineages diverged at least 0.117 Ma but since then have retained ancestral genetic polymorphism and/or experienced gene flow.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Genome sequence, comparative analysis, and population genetics of the domestic horse.
- Record: found
- Abstract: found
- Article: not found
The earliest horse harnessing and milking.
- Record: found
- Abstract: not found
- Article: not found