- Record: found
- Abstract: found
- Article: found
A View of Pre-mRNA Splicing from RNase R Resistant RNAs
Read this article at
Abstract
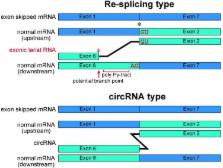
During pre-mRNA splicing, exons in the primary transcript are precisely connected to generate an mRNA. Intron lariat RNAs are formed as by-products of this process. In addition, some exonic circular RNAs (circRNAs) may also result from exon skipping as by-products. Lariat RNAs and circRNAs are both RNase R resistant RNAs. RNase R is a strong 3' to 5' exoribonuclease, which efficiently degrades linear RNAs, such as mRNAs and rRNAs; therefore, the circular parts of lariat RNAs and the circRNAs can be segregated from eukaryotic total RNAs by their RNase R resistance. Thus, RNase R resistant RNAs could provide unexplored splicing information not available from mRNAs. Analyses of these RNAs identified repeating splicing phenomena, such as re-splicing of mature mRNAs and nested splicing. Moreover, circRNA might function as microRNA sponges. There is an enormous variety of endogenous circRNAs, which are generally synthesized in cells and tissues.
Related collections
Most cited references28

- Record: found
- Abstract: not found
- Article: not found
Circular RNAs: splicing's enigma variations.
- Record: found
- Abstract: found
- Article: not found