- Record: found
- Abstract: found
- Article: found
The complete chloroplast genome sequence of Photinia × fraseri, a medicinal plant and phylogenetic analysis
Read this article at
Abstract
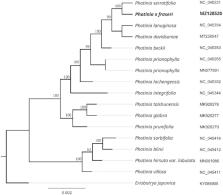
Photinia × fraseri is a common ornamental arbor in the genus Photinia (family Rosaceae), which complete chloroplast (cp) genome was sequenced, assembled and annotated. The chloroplast genome of P. fraseri was 160,184 bp in length, including a large single copy (LSC) region of 88,121, a small single copy (SSC) region of 19,295 bp, and two inverted repeat (IR) regions of 26,384 bp. The GC contents of LSC, SSC, IR and whole genome are 36.5%, 34.1%, 30.3%, and 42.7%, respectively. There are 131 genes annotated, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogenetic analysis revealed that P. fraseri was most related to Photinia serratifolia as a sister group with 100% bootstrap support. The complete chloroplast genome sequences of P. fraseri will provide valuable genomic information to further illuminate phylogenetic classification of Photinia genus.
Related collections
Most cited references10

- Record: found
- Abstract: found
- Article: found
Trimmomatic: a flexible trimmer for Illumina sequence data

- Record: found
- Abstract: found
- Article: found
MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability
- Record: found
- Abstract: found
- Article: not found