- Record: found
- Abstract: found
- Article: found
The double-edged sword of probiotic supplementation on gut microbiota structure in Helicobacter pylori management
Read this article at
ABSTRACT
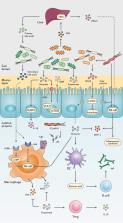
As Helicobacter pylori management has become more challenging and less efficient over the last decade, the interest in innovative interventions is growing by the day. Probiotic co-supplementation to antibiotic therapies is reported in several studies, presenting a moderate reduction in drug-related side effects and a promotion in positive treatment outcomes. However, the significance of gut microbiota involvement in the competence of probiotic co-supplementation is emphasized by a few researchers, indicating the alteration in the host gastrointestinal microbiota following probiotic and drug uptake. Due to the lack of long-term follow-up studies to determine the efficiency of probiotic intervention in H. pylori eradication, and the delicate interaction of the gut microbiota with the host wellness, this review aims to discuss the gut microbiota alteration by probiotic co-supplementation in H. pylori management to predict the comprehensive effectiveness of probiotic oral administration.
Abbreviations: acyl-CoA- acyl-coenzyme A; AMP- antimicrobial peptide; AMPK- AMP-activated protein kinase; AP-1- activator protein 1; BA- bile acid; BAR- bile acid receptor; BCAA- branched-chain amino acid; C2- acetate; C3- propionate; C4- butyrate; C5- valeric acid; CagA- Cytotoxin-associated gene A; cAMP- cyclic adenosine monophosphate; CD- Crohn’s disease; CDI- C. difficile infection; COX-2- cyclooxygenase-2; DC- dendritic cell; EMT- epithelial-mesenchymal transition; FMO- flavin monooxygenases; FXR- farnesoid X receptor; GPBAR1- G-protein-coupled bile acid receptor 1; GPR4- G protein-coupled receptor 4; H2O2- hydrogen peroxide; HCC- hepatocellular carcinoma; HSC- hepatic stellate cell; IBD- inflammatory bowel disease; IBS- irritable bowel syndrome; IFN-γ- interferon-gamma; IgA immunoglobulin A; IL- interleukin; iNOS- induced nitric oxide synthase; JAK1- janus kinase 1; JAM-A- junctional adhesion molecule A; LAB- lactic acid bacteria; LPS- lipopolysaccharide; MALT- mucosa-associated lymphoid tissue; MAMP- microbe-associated molecular pattern; MCP-1- monocyte chemoattractant protein-1; MDR- multiple drug resistance; mTOR- mammalian target of rapamycin; MUC- mucin; NAFLD- nonalcoholic fatty liver disease; NF-κB- nuclear factor kappa B; NK- natural killer; NLRP3- NLR family pyrin domain containing 3; NOC- N-nitroso compounds; NOD- nucleotide-binding oligomerization domain; PICRUSt- phylogenetic investigation of communities by reconstruction of unobserved states; PRR- pattern recognition receptor; RA- retinoic acid; RNS- reactive nitrogen species; ROS- reactive oxygen species; rRNA- ribosomal RNA; SCFA- short-chain fatty acids; SDR- single drug resistance; SIgA- secretory immunoglobulin A; STAT3- signal transducer and activator of transcription 3; T1D- type 1 diabetes; T2D- type 2 diabetes; Th17- T helper 17; TLR- toll-like receptor; TMAO- trimethylamine N-oxide; TML- trimethyllysine; TNF-α- tumor necrosis factor-alpha; Tr1- type 1 regulatory T cell; Treg- regulatory T cell; UC- ulcerative colitis; VacA- Vacuolating toxin A.
Related collections
Most cited references264
- Record: found
- Abstract: found
- Article: not found
Human gut microbiome viewed across age and geography
- Record: found
- Abstract: found
- Article: not found
Gut microbiota in human metabolic health and disease

- Record: found
- Abstract: found
- Article: found