- Record: found
- Abstract: found
- Article: found
A transient ischemic environment induces reversible compaction of chromatin
Read this article at
Abstract
Background
Cells detect and adapt to hypoxic and nutritional stress through immediate transcriptional, translational and metabolic responses. The environmental effects of ischemia on chromatin nanostructure were investigated using single molecule localization microscopy of DNA binding dyes and of acetylated histones, by the sensitivity of chromatin to digestion with DNAseI, and by fluorescence recovery after photobleaching (FRAP) of core and linker histones.
Results
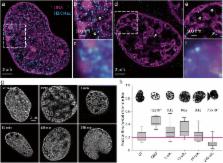
Short-term oxygen and nutrient deprivation of the cardiomyocyte cell line HL-1 induces a previously undescribed chromatin architecture, consisting of large, chromatin-sparse voids interspersed between DNA-dense hollow helicoid structures 40–700 nm in dimension. The chromatin compaction is reversible, and upon restitution of normoxia and nutrients, chromatin transiently adopts a more open structure than in untreated cells. The compacted state of chromatin reduces transcription, while the open chromatin structure induced upon recovery provokes a transitory increase in transcription. Digestion of chromatin with DNAseI confirms that oxygen and nutrient deprivation induces compaction of chromatin. Chromatin compaction is associated with depletion of ATP and redistribution of the polyamine pool into the nucleus. FRAP demonstrates that core histones are not displaced from compacted chromatin; however, the mobility of linker histone H1 is considerably reduced, to an extent that far exceeds the difference in histone H1 mobility between heterochromatin and euchromatin.
Related collections
Most cited references83
- Record: found
- Abstract: found
- Article: not found
A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping.
- Record: found
- Abstract: found
- Article: not found
Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions
- Record: found
- Abstract: found
- Article: not found