- Record: found
- Abstract: found
- Article: found
Properties and Modeling of GWAS when Complex Disease Risk Is Due to Non-Complementing, Deleterious Mutations in Genes of Large Effect
Read this article at
Abstract
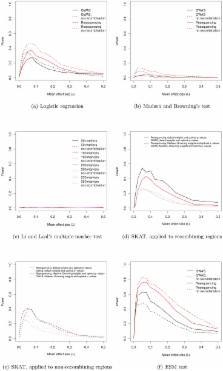
Current genome-wide association studies (GWAS) have high power to detect intermediate frequency SNPs making modest contributions to complex disease, but they are underpowered to detect rare alleles of large effect (RALE). This has led to speculation that the bulk of variation for most complex diseases is due to RALE. One concern with existing models of RALE is that they do not make explicit assumptions about the evolution of a phenotype and its molecular basis. Rather, much of the existing literature relies on arbitrary mapping of phenotypes onto genotypes obtained either from standard population-genetic simulation tools or from non-genetic models. We introduce a novel simulation of a 100-kilobase gene region, based on the standard definition of a gene, in which mutations are unconditionally deleterious, are continuously arising, have partially recessive and non-complementing effects on phenotype (analogous to what is widely observed for most Mendelian disorders), and are interspersed with neutral markers that can be genotyped. Genes evolving according to this model exhibit a characteristic GWAS signature consisting of an excess of marginally significant markers. Existing tests for an excess burden of rare alleles in cases have low power while a simple new statistic has high power to identify disease genes evolving under our model. The structure of linkage disequilibrium between causative mutations and significantly associated markers under our model differs fundamentally from that seen when rare causative markers are assumed to be neutral. Rather than tagging single haplotypes bearing a large number of rare causative alleles, we find that significant SNPs in a GWAS tend to tag single causative mutations of small effect relative to other mutations in the same gene. Our results emphasize the importance of evaluating the power to detect associations under models that are genetically and evolutionarily motivated.
Author Summary
Current GWA studies typically only explain a small fraction of heritable variation in complex traits, resulting in speculation that a large fraction of variation in such traits may be due to rare alleles of large effect (RALE). The most parsimonious evolutionary mechanism that results in an inverse relationship between the frequency and effect size of causative alleles is an equilibrium between newly arising deleterious mutations and selection eliminating those mutations, resulting in an inverse relation between effect size and average frequency. This assumption is not built into many current models of RALE and, as a result, power calculations may be misleading. We use forward population genetic simulations to explore the ability of GWAS to detect genes in which unconditionally deleterious, partially recessive mutations arise each generation. Our model is based on the standard definition of a gene as a region within which loss-of-function mutations fail to complement, consistent with the multi-allelic basis for Mendelian disorders. Our model predicts that it may not be uncommon for single genes evolving under our model to contribute upwards of 5% to variation in a complex trait, and that such genes could be routinely detected via modified GWAS approaches.