- Record: found
- Abstract: found
- Article: found
Holliday junction resolution: Regulation in space and time
Read this article at
Abstract
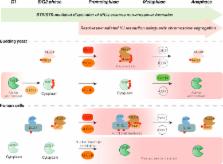
Holliday junctions (HJs) can be formed between sister chromatids or homologous chromosomes during the recombinational repair of DNA lesions. A variety of pathways act upon HJs to remove them from DNA, in events that are critical for appropriate chromosome segregation. Despite the identification and characterization of multiple enzymes involved in HJ processing, the cellular mechanisms that regulate and implement pathway usage have only just started to be delineated. A conserved network of core cell-cycle kinases and phosphatases modulate HJ metabolism by exerting spatial and temporal control over the activities of two structure-selective nucleases: yeast Mus81-Mms4 (human MUS81-EME1) and Yen1 (human GEN1). These regulatory cycles operate to establish the sequential activation of HJ processing enzymes, implementing a hierarchy in pathway usage that ensure the elimination of chromosomal interactions which would otherwise interfere with chromosome segregation. Mus81-Mms4/EME1 and Yen1/GEN1 emerge to define a special class of enzymes, evolved to satisfy the cellular need of safeguarding the completion of DNA repair when on the verge of chromosome segregation.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
The Bloom's syndrome helicase suppresses crossing over during homologous recombination.
- Record: found
- Abstract: found
- Article: not found
The Bloom's syndrome gene product is homologous to RecQ helicases.
- Record: found
- Abstract: found
- Article: not found