- Record: found
- Abstract: found
- Article: found
The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver
Read this article at
Abstract
Background
Geese were domesticated over 6,000 years ago, making them one of the first domesticated poultry. Geese are capable of rapid growth, disease resistance, and high liver lipid storage capacity, and can be easily fed coarse fodder. Here, we sequence and analyze the whole-genome sequence of an economically important goose breed in China and compare it with that of terrestrial bird species.
Results
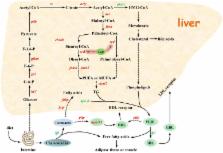
A draft sequence of the whole-goose genome was obtained by shotgun sequencing, and 16,150 protein-coding genes were predicted. Comparative genomics indicate that significant differences occur between the goose genome and that of other terrestrial bird species, particularly regarding major histocompatibility complex, Myxovirus resistance, Retinoic acid-inducible gene I, and other genes related to disease resistance in geese. In addition, analysis of transcriptome data further reveals a potential molecular mechanism involved in the susceptibility of geese to fatty liver disease and its associated symptoms, including high levels of unsaturated fatty acids and low levels of cholesterol. The results of this study show that deletion of the goose lep gene might be the result of positive selection, thus allowing the liver to adopt energy storage mechanisms for long-distance migration.
Related collections
Most cited references43
- Record: found
- Abstract: found
- Article: not found
MRBAYES: Bayesian inference of phylogenetic trees.
- Record: found
- Abstract: found
- Article: not found
RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates.
- Record: found
- Abstract: found
- Article: not found