- Record: found
- Abstract: found
- Article: found
The temporal dynamics of chromosome instability in ovarian cancer cell lines and primary patient samples
Read this article at
Abstract
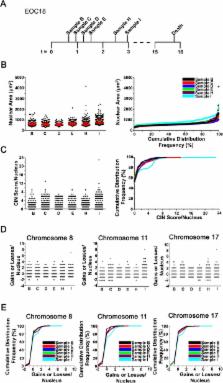
Epithelial ovarian cancer (EOC) is the most prevalent form of ovarian cancer and has the highest mortality rate. Novel insight into EOC is required to minimize the morbidity and mortality rates caused by recurrent, drug resistant disease. Although numerous studies have evaluated genome instability in EOC, none have addressed the putative role chromosome instability (CIN) has in disease progression and drug resistance. CIN is defined as an increase in the rate at which whole chromosomes or large parts thereof are gained or lost, and can only be evaluated using approaches capable of characterizing genetic or chromosomal heterogeneity within populations of cells. Although CIN is associated with numerous cancer types, its prevalence and dynamics in EOC is unknown. In this study, we assessed CIN within serial samples collected from the ascites of five EOC patients, and in two well-established ovarian cancer cell models of drug resistance (PEO1/4 and A2780s/cp). We quantified and compared CIN (as measured by nuclear areas and CIN Score (CS) values) within and between serial samples to glean insight into the association and dynamics of CIN within EOC, with a particular focus on resistant and recurrent disease. Using quantitative, single cell analyses we determined that CIN is associated with every sample evaluated and further show that many EOC samples exhibit a large degree of nuclear size and CS value heterogeneity. We also show that CIN is dynamic and generally increases within resistant disease. Finally, we show that both drug resistance models (PEO1/4 and A2780s/cp) exhibit heterogeneity, albeit to a much lesser extent. Surprisingly, the two cell line models exhibit remarkably similar levels of CIN, as the nuclear areas and CS values are largely overlapping between the corresponding paired lines. Accordingly, these data suggest CIN may represent a novel biomarker capable of monitoring changes in EOC progression associated with drug resistance.
Author summary
Ovarian cancer is one of the most lethal cancers in women due to the high prevalence of drug resistant disease. New insight into the biology causing drug resistance is required to reduce death rates associated with the disease. In many cancer types, chromosome instability (CIN; or abnormal numbers of chromosomes) is associated with aggressive tumours, the acquisition of multi-drug resistance and poor patient outcome, yet CIN is poorly characterized in ovarian cancer. Here, we employ a new microscopy-based approach to examine the presence and dynamics of CIN within single cells obtained from ovarian cancer patients collected over time. We show that CIN is associated with every sample and further show that increases in CIN are associated with treatment and/or the development of drug (platinum) resistant disease. Although additional studies are required, these findings suggest CIN may be a new biomarker that can monitor disease progression, particularly in response to therapy. Finally, we demonstrate that CIN is low in two cellular models of drug resistant disease. Collectively, our findings strengthen the need for studies employing patient-derived samples to accurately assess the abnormal biology associated with the disease, and cautions the use of these cell models in studies related to drug resistance that may arise from CIN.
Related collections
Most cited references41
- Record: found
- Abstract: found
- Article: not found
Genetic instabilities in human cancers.
- Record: found
- Abstract: found
- Article: not found
Mechanisms of chromosomal instability.
- Record: found
- Abstract: found
- Article: found