- Record: found
- Abstract: found
- Article: found
Genome-Wide Characterization of the HSP20 Gene Family Identifies Potential Members Involved in Temperature Stress Response in Apple
Read this article at
Abstract
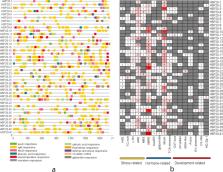
Apple ( Malus domestica Borkh.), an economically important tree fruit worldwide, frequently suffers from temperature stress during growth and development, which strongly affects the yield and quality. Heat shock protein 20 ( HSP20) genes play crucial roles in protecting plants against abiotic stresses. However, they have not been systematically investigated in apple. In this study, we identified 41 HSP20 genes in the apple ‘Golden Delicious’ genome. These genes were unequally distributed on 15 different chromosomes and were classified into 10 subfamilies based on phylogenetic analysis and predicted subcellular localization. Chromosome mapping and synteny analysis indicated that three pairs of apple HSP20 genes were tandemly duplicated. Sequence analysis revealed that all apple HSP20 proteins reflected high structure conservation and most apple HSP20 genes (92.6%) possessed no introns, or only one intron. Numerous apple HSP20 gene promoter sequences contained stress and hormone response cis-elements. Transcriptome analysis revealed that 35 of 41 apple HSP20 genes were nearly unchanged or downregulated under normal temperature and cold stress, whereas these genes exhibited high-expression levels under heat stress. Subsequent qRT-PCR results showed that 12 of 29 selected apple HSP20 genes were extremely up-regulated (more than 1,000-fold) after 4 h of heat stress. However, the heat-upregulated genes were barely expressed or downregulated in response to cold stress, which indicated their potential function in mediating the response of apple to heat stress. Taken together, these findings lay the foundation to functionally characterize HSP20 genes to unravel their exact role in heat defense response in apple.
Related collections
Most cited references40
- Record: found
- Abstract: found
- Article: not found
Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
- Record: found
- Abstract: found
- Article: not found
HISAT: a fast spliced aligner with low memory requirements.
- Record: found
- Abstract: found
- Article: not found