- Record: found
- Abstract: found
- Article: found
Alternative proteins are functional regulators in cell reprogramming by PKA activation
Read this article at
Abstract
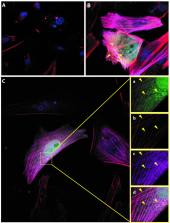
It has been recently shown that many proteins are lacking from reference databases used in mass spectrometry analysis, due to their translation templated on alternative open reading frames. This questions our current understanding of gene annotation and drastically expands the theoretical proteome complexity. The functions of these alternative proteins (AltProts) still remain largely unknown. We have developed a large-scale and unsupervised approach based on cross-linking mass spectrometry (XL-MS) followed by shotgun proteomics to gather information on the functional role of AltProts by mapping them back into known signalling pathways through the identification of their reference protein (RefProt) interactors. We have identified and profiled AltProts in a cancer cell reprogramming system: NCH82 human glioma cells after 0, 16, 24 and 48 h Forskolin stimulation. Forskolin is a protein kinase A activator inducing cell differentiation and epithelial–mesenchymal transition. Our data show that AltMAP2, AltTRNAU1AP and AltEPHA5 interactions with tropomyosin 4 are downregulated under Forskolin treatment. In a wider perspective, Gene Ontology and pathway enrichment analysis (STRING) revealed that RefProts associated with AltProts are enriched in cellular mobility and transfer RNA regulation. This study strongly suggests novel roles of AltProts in multiple essential cellular functions and supports the importance of considering them in future biological studies.
Related collections
Most cited references74
- Record: found
- Abstract: found
- Article: not found
Cytoscape: a software environment for integrated models of biomolecular interaction networks.
- Record: found
- Abstract: found
- Article: not found
UCSF Chimera--a visualization system for exploratory research and analysis.

- Record: found
- Abstract: found
- Article: found