- Record: found
- Abstract: found
- Article: found
Clinical, Genomic, and Transcriptomic Data Profiling of Biliary Tract Cancer Reveals Subtype-Specific Immune Signatures

Read this article at
PURPOSE
Biliary tract cancers (BTCs) are aggressive cancers that carry a poor prognosis. An enhanced understanding of the immune landscape of anatomically and molecularly defined subsets of BTC may improve patient selection for immunotherapy and inform immune-based combination treatment strategies.
METHODS
We analyzed deidentified clinical, genomic, and transcriptomic data from the Tempus database to determine the mutational frequency and mutational clustering across the three major BTC subtypes (intrahepatic cholangiocarcinoma [IHC], extrahepatic cholangiocarcinoma, and gallbladder cancer). We subsequently determined the relationship between specific molecular alterations and anatomical subsets and features of the BTC immune microenvironment.
RESULTS
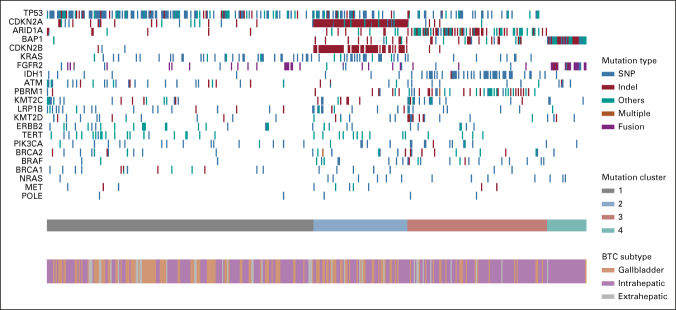
We analyzed 454 samples of BTC, of which the most commonly detected alterations were TP53 (42.5%), CDKN2A (23.4%), ARID1A (19.6%), BAP1 (15.5%), KRAS (15%), CDKN2B (14.2%), PBRM1 (11.7%), IDH1 (11.7%), TERT (8.4%), KMT2C (10.4%) and LRP1B (8.4%), and FGFR2 fusions (8.7%). Potentially actionable molecular alterations were identified in 30.5% of BTCs including 39.1% of IHC. Integrative cluster analysis revealed four distinct molecular clusters, with cluster 4 predominately associated with FGFR2 rearrangements and BAP1 mutations in IHC. Immune-related biomarkers indicative of an inflamed tumor-immune microenvironment were elevated in gallbladder cancers and in cluster 1, which was enriched for TP53, KRAS, and ATM mutations. Multiple common driver genes, including TP53, FGFR2, IDH1, TERT, BRAF, and BAP1, were individually associated with unique BTC immune microenvironments.
Abstract
Related collections
Most cited references35
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found
featureCounts: an efficient general purpose program for assigning sequence reads to genomic features.
- Record: found
- Abstract: found
- Article: not found