- Record: found
- Abstract: found
- Article: found
The SET1 Complex Selects Actively Transcribed Target Genes via Multivalent Interaction with CpG Island Chromatin

Read this article at
Summary
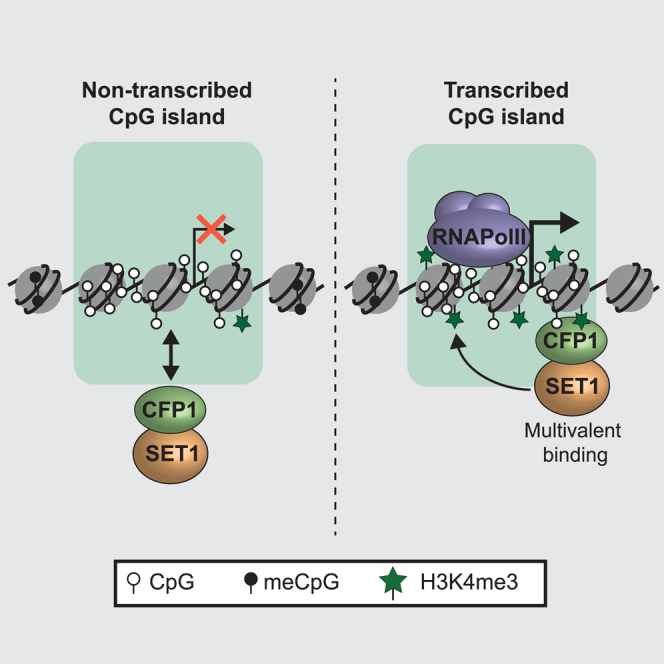
Chromatin modifications and the promoter-associated epigenome are important for the regulation of gene expression. However, the mechanisms by which chromatin-modifying complexes are targeted to the appropriate gene promoters in vertebrates and how they influence gene expression have remained poorly defined. Here, using a combination of live-cell imaging and functional genomics, we discover that the vertebrate SET1 complex is targeted to actively transcribed gene promoters through CFP1, which engages in a form of multivalent chromatin reading that involves recognition of non-methylated DNA and histone H3 lysine 4 trimethylation (H3K4me3). CFP1 defines SET1 complex occupancy on chromatin, and its multivalent interactions are required for the SET1 complex to place H3K4me3. In the absence of CFP1, gene expression is perturbed, suggesting that normal targeting and function of the SET1 complex are central to creating an appropriately functioning vertebrate promoter-associated epigenome.
Graphical Abstract
Highlights
-
•
The CFP1/SET1 complex engages in dynamic and stable chromatin-binding events
-
•
CFP1 uses multivalent chromatin interactions to select active CpG island promoters
-
•
SET1A occupancy at CpG island promoters is predominately defined by CFP1
-
•
CFP1 targets SET1 to shape promoter-associated H3K4me3 and gene expression
Abstract
Brown et al. show that the SET1 complex is driven to active CpG island promoters via the CFP1 protein, which engages in multivalent chromatin binding to recognize both non-methylated DNA and H3K4me3. This is necessary for normal H3K4me3 at active gene promoters and appropriate regulation of gene expression.
Related collections
Most cited references52
- Record: found
- Abstract: found
- Article: not found
The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis.
- Record: found
- Abstract: found
- Article: not found
Histone exchange, chromatin structure and the regulation of transcription.
- Record: found
- Abstract: found
- Article: not found