- Record: found
- Abstract: found
- Article: not found
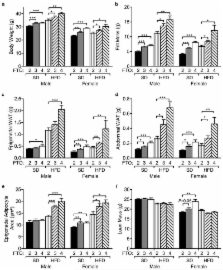
Overexpression of Fto leads to increased food intake and results in obesity
Abstract
Genome-wide association studies have identified SNPs within the human FTO gene that display a strong association with obesity. Individuals homozygous for the at-risk rs9939609 A allele weigh ~3kg more. Loss of function and/or expression of FTO in mice leads to increased energy expenditure and a lean phenotype. We show here that ubiquitous overexpression of Fto leads to a dose-dependent increase in body and fat mass, irrespective of whether mice are fed a standard or high fat diet. The increased body mass results primarily from increased food intake. Glucose intolerance develops with increased Fto expression on a high fat diet. This study provides the first direct evidence that increased Fto expression causes obesity in mice.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase.
- Record: found
- Abstract: found
- Article: not found
Inactivation of the Fto gene protects from obesity.
- Record: found
- Abstract: found
- Article: not found
