- Record: found
- Abstract: found
- Article: found
Defining Epidermal Basal Cell States during Skin Homeostasis and Wound Healing Using Single-Cell Transcriptomics
Read this article at
SUMMARY
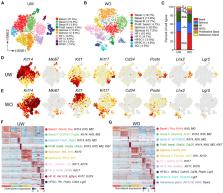
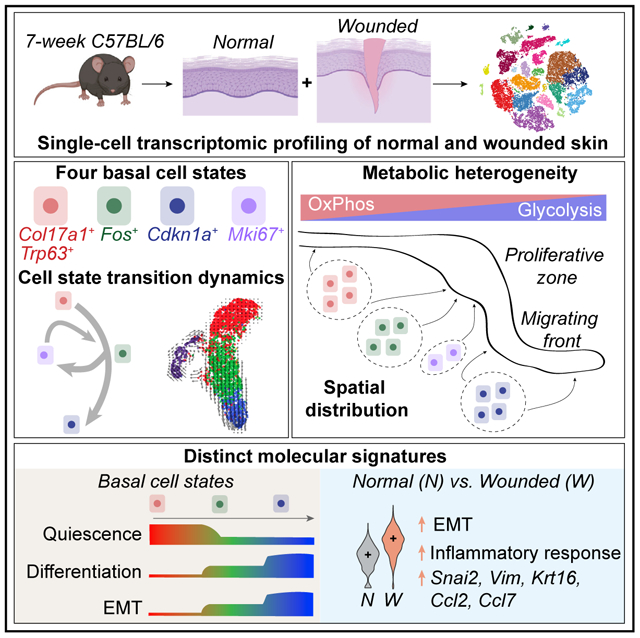
Our knowledge of transcriptional heterogeneities in epithelial stem and progenitor cell compartments is limited. Epidermal basal cells sustain cutaneous tissue maintenance and drive wound healing. Previous studies have probed basal cell heterogeneity in stem and progenitor potential, but a comprehensive dissection of basal cell dynamics during differentiation is lacking. Using single-cell RNA sequencing coupled with RNAScope and fluorescence lifetime imaging, we identify three non-proliferative and one proliferative basal cell state in homeostatic skin that differ in metabolic preference and become spatially partitioned during wound re-epithelialization. Pseudotemporal trajectory and RNA velocity analyses predict a quasi-linear differentiation hierarchy where basal cells progress from Col17a1 Hi /Trp63 Hi state to early-response state, proliferate at the juncture of these two states, or become growth arrested before differentiating into spinous cells. Wound healing induces plasticity manifested by dynamic basal-spinous interconversions at multiple basal transcriptional states. Our study provides a systematic view of epidermal cellular dynamics, supporting a revised “hierarchical-lineage” model of homeostasis.
Graphical Abstract
In Brief
Haensel et al. performed a comprehensive dissection of the cellular makeup of skin during homeostasis and wound healing and the molecular heterogeneity and cellular dynamics within its stem-cell-containing epidermal basal layer. Their work provides insights and stimulates further investigation into the mechanism of skin maintenance and repair.
Related collections
Most cited references61
- Record: found
- Abstract: found
- Article: not found
p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development.
- Record: found
- Abstract: found
- Article: not found
Molecular regulation of stem cell quiescence.
- Record: found
- Abstract: found
- Article: not found