- Record: found
- Abstract: found
- Article: found
Meiotic prophase roles of Rec8 in crossover recombination and chromosome structure
Read this article at
Abstract
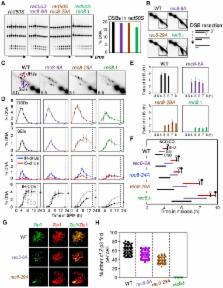
Rec8 is a prominent component of the meiotic prophase chromosome axis that mediates sister chromatid cohesion, homologous recombination and chromosome synapsis. Here, we explore the prophase roles of Rec8. (i) During the meiotic divisions, Rec8 phosphorylation mediates its separase-mediated cleavage. We show here that such cleavage plays no detectable role for chromosomal events of prophase. (ii) We have analyzed in detail three rec8 phospho-mutants, with 6, 24 or 29 alanine substitutions. A distinct ‘separation of function’ phenotype is revealed. In the mutants, axis formation and recombination initiation are normal, as is non-crossover recombination; in contrast, crossover (CO)-related events are defective. Moreover, the severities of these defects increase coordinately with the number of substitution mutations, consistent with the possibility that global phosphorylation of Rec8 is important for these effects. (iii) We have analyzed the roles of three kinases that phosphorylate Rec8 during prophase. Timed inhibition of Dbf4-dependent Cdc7 kinase confers defects concordant with rec8 phospho-mutant phenotypes. Inhibition of Hrr25 or Cdc5/polo-like kinase does not. Our results suggest that Rec8's prophase function, independently of cohesin cleavage, contributes to CO-specific events in conjunction with the maintenance of homolog bias at the leptotene/zygotene transition of meiotic prophase.
Related collections
Most cited references46
- Record: found
- Abstract: found
- Article: not found
A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis.
- Record: found
- Abstract: found
- Article: not found
The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination.
- Record: found
- Abstract: found
- Article: not found