- Record: found
- Abstract: found
- Article: found
DNA methylation: a potential mediator between air pollution and metabolic syndrome
Read this article at
Abstract
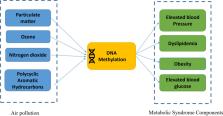
Given the global increase in air pollution and its crucial role in human health, as well as the steep rise in prevalence of metabolic syndrome (MetS), a better understanding of the underlying mechanisms by which environmental pollution may influence MetS is imperative. Exposure to air pollution is known to impact DNA methylation, which in turn may affect human health. This paper comprehensively reviews the evidence for the hypothesis that the effect of air pollution on the MetS is mediated by DNA methylation in blood. First, we present a summary of the impact of air pollution on metabolic dysregulation, including the components of MetS, i.e., disorders in blood glucose, lipid profile, blood pressure, and obesity. Then, we provide evidence on the relation between air pollution and endothelial dysfunction as one possible mechanism underlying the relation between air pollution and MetS. Subsequently, we review the evidence that air pollution (PM, ozone, NO 2 and PAHs) influences DNA methylation. Finally, we summarize association studies between DNA methylation and MetS. Integration of current evidence supports our hypothesis that methylation may partly mediate the effect of air pollution on MetS.
Related collections
Most cited references133

- Record: found
- Abstract: found
- Article: found
Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the Global Burden of Diseases Study 2015
- Record: found
- Abstract: found
- Article: not found
The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses.
- Record: found
- Abstract: found
- Article: not found