- Record: found
- Abstract: found
- Article: found
Expression of human eukaryotic initiation factor 3f oscillates with cell cycle in A549 cells and is essential for cell viability
Read this article at
Abstract
Background
Transcriptional and postranslational regulation of the cell cycle has been widely studied. However, there is scarce knowledge concerning translational control of this process. Several mammalian eukaryotic initiation factors (eIFs) seem to be implicated in controlling cell proliferation. In this work, we investigated if the human eIF3f expression and function is cell cycle related.
Results
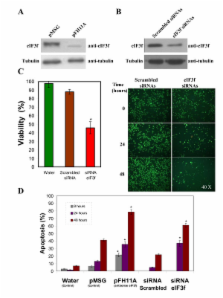
The human eIF3f expression has been found to be upregulated in growth-stimulated A549 cells and downregulated in G0. Western blot analysis and eIF3f promotor-luciferase fusions revealed that eIF3f expression peaks twice in the cell cycle: in the S and the M phases. Deregulation of eIF3f expression negatively affects cell viability and induces apoptosis.
Conclusions
The expression pattern of human eIF3f during the cell cycle confirms that this gene is cell division related. The fact that eIF3f expression peaks in two cell cycle phases raises the possibility that this gene may exert a differential function in the S and M phases. Our results strongly suggest that eIF3f is essential for cell proliferation.
Related collections
Most cited references29
- Record: found
- Abstract: found
- Article: not found
Evolutionary fate of retroposed gene copies in the human genome.
- Record: found
- Abstract: found
- Article: not found
Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity.
- Record: found
- Abstract: found
- Article: not found