- Record: found
- Abstract: found
- Article: found
A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer
Read this article at
Abstract
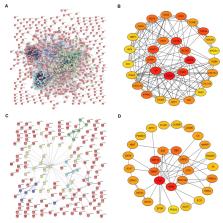
Background: Recently, increasing evidence has uncovered the roles of mRNA-miRNA-lncRNA network in multiple human cancers. However, a systematic mRNA-miRNA-lncRNA network linked to pancreatic cancer prognosis is still absent. Methods: Differentially expressed genes (DEGs) were first identified by mining GSE16515 and GSE15471 datasets. DAVID database was utilized to conduct functional enrichment analysis. Protein-protein interaction (PPI) network was built using STRING database, and hub genes were identified by Cytoscape plug-in CytoHubba. Upstream miRNAs and lncRNAs of mRNAs were predicted by miRTarBase and miRNet, respectively. Expression, survival and correlation analysis for genes, miRNAs and lncRNAs were performed via GEPIA, Kaplan-Meier plotter and starBase. Results: 734 and 180 upregulated and downregulated significant DEGs were identified, respectively. Functional enrichment analysis revealed that they were significantly enriched in focal adhesion, pathways in cancer and metabolic pathways. According to node degree, hub genes in the PPI networks were screened, such as TGFB1 and ALB. Among the top 20 hub genes, 7 upregulated genes and 2 downregulated hub genes had significant prognostic values in pancreatic cancer. 33 miRNAs were predicted to target the 9 key genes. But only high expression of 8 miRNAs indicated favorable prognosis in pancreatic cancer. Then, 90 lncRNAs were predicted to potentially bind to the 8 miRNAs. SCAMP1, HCP5, MAL2 and LINC00511 were finally identified as key lncRNAs. By combination of results from expression, survival and correlation analysis demonstrated that MMP9/ITGB1-miR-29b-3p-HCP5 competing endogenous RNA (ceRNA) sub-network was linked to prognosis of pancreatic cancer. Conclusions: In a word, we established a novel mRNA-miRNA-lncRNA sub-network, among which each RNA may be utilized as a prognostic biomarker of pancreatic cancer.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Gene Ontology: tool for the unification of biology
- Record: found
- Abstract: found
- Article: not found
miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients.
- Record: found
- Abstract: found
- Article: not found