- Record: found
- Abstract: found
- Article: found
Autistic traits in myotonic dystrophy type 1 due to MBNL inhibition and RNA mis-splicing
Read this article at
Abstract
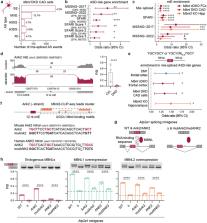
Tandem repeat expansions are enriched in autism spectrum disorder, including CTG expansion in the DMPK gene that underlines myotonic muscular dystrophy type 1. Although the clinical connection of autism to myotonic dystrophy is corroborated, the molecular links remained unknown. Here, we show a mechanistic path of autism via repeat expansion in myotonic dystrophy. We found that inhibition of muscleblind-like (MBNL) splicing factors by expanded CUG RNAs alerts the splicing of autism-risk genes during brain development especially a class of autism-relevant microexons. To provide in vivo evidence that the CTG expansion and MBNL inhibition axis leads to the presentation of autistic traits, we demonstrate that CTG expansion and MBNL-null mouse models recapitulate autism-relevant mis-splicing profiles and demonstrate social deficits. Our findings indicate that DMPK CTG expansion-associated autism arises from developmental mis-splicing. Understanding this pathomechanistic connection provides an opportunity for greater in-depth investigations of mechanistic threads in autism.
Related collections
Most cited references84

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
STAR: ultrafast universal RNA-seq aligner.
- Record: found
- Abstract: found
- Article: not found