- Record: found
- Abstract: found
- Article: found
Six Homeoproteins Directly Activate Myod Expression in the Gene Regulatory Networks That Control Early Myogenesis
Read this article at
Abstract
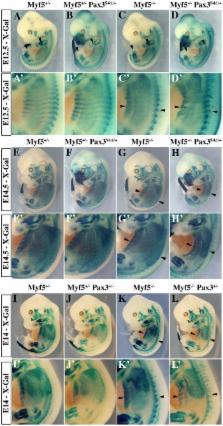
In mammals, several genetic pathways have been characterized that govern engagement of multipotent embryonic progenitors into the myogenic program through the control of the key myogenic regulatory gene Myod. Here we demonstrate the involvement of Six homeoproteins. We first targeted into a Pax3 allele a sequence encoding a negative form of Six4 that binds DNA but cannot interact with essential Eya co-factors. The resulting embryos present hypoplasic skeletal muscles and impaired Myod activation in the trunk in the absence of Myf5/Mrf4. At the axial level, we further show that Myod is still expressed in compound Six1/Six4:Pax3 but not in Six1/Six4:Myf5 triple mutant embryos, demonstrating that Six1/4 participates in the Pax3-Myod genetic pathway. Myod expression and head myogenesis is preserved in Six1/Six4:Myf5 triple mutant embryos, illustrating that upstream regulators of Myod in different embryonic territories are distinct. We show that Myod regulatory regions are directly controlled by Six proteins and that, in the absence of Six1 and Six4, Six2 can compensate.
Author Summary
The onset of skeletal muscle formation is controlled by complex gene regulatory networks. By manipulation of these genetic pathways in the mouse embryo, we have examined the interplay between genes encoding the transcriptional regulator Pax3; the major myogenic determination proteins Myf5, Mrf4, and Myod; as well as genes encoding homeodomain proteins Six1 and Six4. In the absence of Myf5 and Six1/4, Myod expression is compromised. We demonstrate that key regulatory elements of the Myod gene are directly targeted by Six factors, including Six2, which is unexpectedly upregulated in the absence of Six1 and Six4. This work therefore reveals new aspects of the gene regulatory networks that control myogenesis.
Related collections
Most cited references37
- Record: found
- Abstract: found
- Article: not found
Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice.
- Record: found
- Abstract: found
- Article: not found
Pax 6: mastering eye morphogenesis and eye evolution.
- Record: found
- Abstract: found
- Article: not found