- Record: found
- Abstract: found
- Article: found
The Integrator Complex Subunit 6 (Ints6) Confines the Dorsal Organizer in Vertebrate Embryogenesis
Read this article at
Abstract
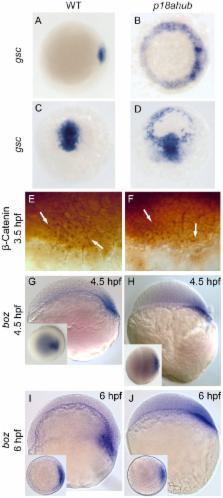
Dorsoventral patterning of the embryonic axis relies upon the mutual antagonism of competing signaling pathways to establish a balance between ventralizing BMP signaling and dorsal cell fate specification mediated by the organizer. In zebrafish, the initial embryo-wide domain of BMP signaling is refined into a morphogenetic gradient following activation dorsally of a maternal Wnt pathway. The accumulation of β-catenin in nuclei on the dorsal side of the embryo then leads to repression of BMP signaling dorsally and the induction of dorsal cell fates mediated by Nodal and FGF signaling. A separate Wnt pathway operates zygotically via Wnt8a to limit dorsal cell fate specification and maintain the expression of ventralizing genes in ventrolateral domains. We have isolated a recessive dorsalizing maternal-effect mutation disrupting the gene encoding Integrator Complex Subunit 6 (Ints6). Due to widespread de-repression of dorsal organizer genes, embryos from mutant mothers fail to maintain expression of BMP ligands, fail to fully express vox and ved, two mediators of Wnt8a, display delayed cell movements during gastrulation, and severe dorsalization. Consistent with radial dorsalization, affected embryos display multiple independent axial domains along with ectopic dorsal forerunner cells. Limiting Nodal signaling or restoring BMP signaling restores wild-type patterning to affected embryos. Our results are consistent with a novel role for Ints6 in restricting the vertebrate organizer to a dorsal domain in embryonic patterning.
Author Summary
A complex integration of signaling pathways establishes the body plan of the vertebrate embryo. The dorsal side of the embryo is defined by the organizer, a specialized field of cells that breaks the symmetry of the zebrafish blastula by instructing neighboring cells to adopt dorsal fates based on their proximity. The isolation of mutant genes in the zebrafish has identified many genes required for embryonic development. However, our knowledge of the molecular mechanisms integrating different signaling pathways within a gene regulatory network to properly pattern the embryo is still incomplete. We isolated a recessive maternal effect mutation in the integrator complex subunit 6 ( ints6) gene that leads to a circumferential expansion of the organizer and the formation of dorsal tissues at the expense of ventral tissues. Currently, the only reported role for the Integrator Complex is to mediate processing of snRNAs of the spliceosome. Our molecular genetic approach indicates that ints6 confines the organizer to dorsal domains, preventing it from extending around the margin into ventral domains. Thus, we have determined a novel role for a highly conserved component of an RNA processing machine.
Related collections
Most cited references92
- Record: found
- Abstract: found
- Article: not found
Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II.
- Record: found
- Abstract: found
- Article: not found
The EGF-CFC protein one-eyed pinhead is essential for nodal signaling.
- Record: found
- Abstract: found
- Article: not found
