- Record: found
- Abstract: found
- Article: found
Disease-associated DNA2 nuclease–helicase protects cells from lethal chromosome under-replication
Read this article at
Abstract
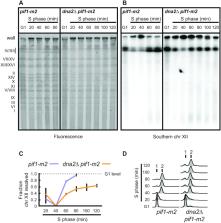
DNA2 is an essential nuclease–helicase implicated in DNA repair, lagging-strand DNA synthesis, and the recovery of stalled DNA replication forks (RFs). In Saccharomyces cerevisiae, dna2Δ inviability is reversed by deletion of the conserved helicase PIF1 and/or DNA damage checkpoint-mediator RAD9. It has been suggested that Pif1 drives the formation of long 5′-flaps during Okazaki fragment maturation, and that the essential function of Dna2 is to remove these intermediates. In the absence of Dna2, 5′-flaps are thought to accumulate on the lagging strand, resulting in DNA damage-checkpoint arrest and cell death. In line with Dna2’s role in RF recovery, we find that the loss of Dna2 results in severe chromosome under-replication downstream of endogenous and exogenous RF-stalling. Importantly, unfaithful chromosome replication in Dna2-mutant cells is exacerbated by Pif1, which triggers the DNA damage checkpoint along a pathway involving Pif1’s ability to promote homologous recombination-coupled replication. We propose that Dna2 fulfils its essential function by promoting RF recovery, facilitating replication completion while suppressing excessive RF restart by recombination-dependent replication (RDR) and checkpoint activation. The critical nature of Dna2’s role in controlling the fate of stalled RFs provides a framework to rationalize the involvement of DNA2 in Seckel syndrome and cancer.
Related collections
Most cited references71
- Record: found
- Abstract: found
- Article: not found
Replication fork reversal in eukaryotes: from dead end to dynamic response.
- Record: found
- Abstract: found
- Article: not found
A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae.
- Record: found
- Abstract: found
- Article: not found