- Record: found
- Abstract: found
- Article: found
Exome sequencing and complex disease: practical aspects of rare variant association studies
Read this article at
Abstract
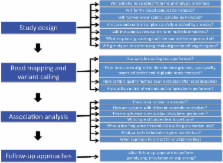
Genetic association and linkage studies can provide insights into complex disease biology, guiding the development of new diagnostic and therapeutic strategies. Over the past decade, genetic association studies have largely focused on common, easy to measure genetic variants shared between many individuals. These common variants typically have subtle functional consequence and translating the resulting association signals into biological insights can be challenging. In the last few years, exome sequencing has emerged as a cost-effective strategy for extending these studies to include rare coding variants, which often have more marked functional consequences. Here, we provide practical guidance in the design and analysis of complex trait association studies focused on rare, coding variants.
Related collections
Most cited references45
- Record: found
- Abstract: found
- Article: not found
Genome-wide association studies for complex traits: consensus, uncertainty and challenges.
- Record: found
- Abstract: found
- Article: not found
Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility.
- Record: found
- Abstract: found
- Article: not found