- Record: found
- Abstract: found
- Article: found
Target-Specific Precision of CRISPR-Mediated Genome Editing

Read this article at
Summary
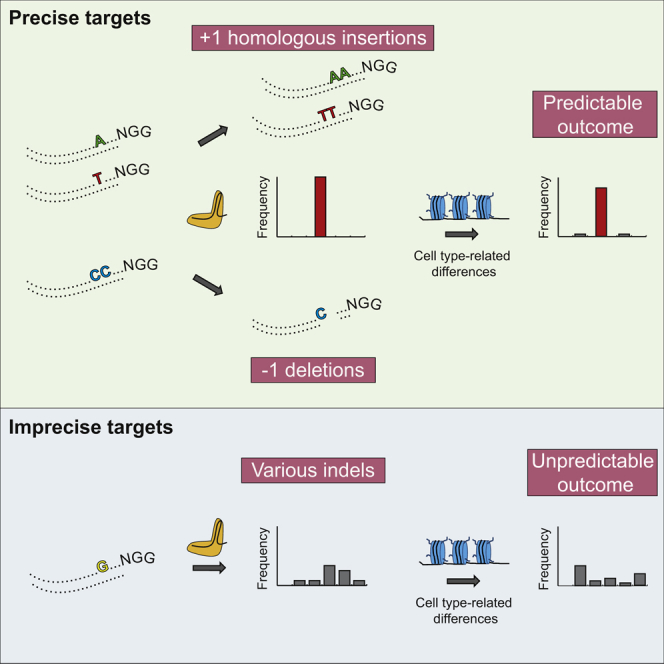
The CRISPR-Cas9 system has successfully been adapted to edit the genome of various organisms. However, our ability to predict the editing outcome at specific sites is limited. Here, we examined indel profiles at over 1,000 genomic sites in human cells and uncovered general principles guiding CRISPR-mediated DNA editing. We find that precision of DNA editing (i.e., recurrence of a specific indel) varies considerably among sites, with some targets showing one highly preferred indel and others displaying numerous infrequent indels. Editing precision correlates with editing efficiency and a preference for single-nucleotide homologous insertions. Precise targets and editing outcome can be predicted based on simple rules that mainly depend on the fourth nucleotide upstream of the protospacer adjacent motif (PAM). Indel profiles are robust, but they can be influenced by chromatin features. Our findings have important implications for clinical applications of CRISPR technology and reveal general patterns of broken end joining that can provide insights into DNA repair mechanisms.
Graphical Abstract
Highlights
Abstract
Chakrabarti, Henser-Brownhill, Monserrat et al. show that the genome-editing outcome can be predicted based on simple rules that mainly depend on the target site sequence. Since editing precision varies considerably across sites, careful selection of a predictable target is critical to induce a desired modification in a cell-type-independent manner.
Related collections
Most cited references12
- Record: found
- Abstract: found
- Article: not found
Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements
- Record: found
- Abstract: found
- Article: not found
Predicting the mutations generated by repair of Cas9-induced double-strand breaks
- Record: found
- Abstract: not found
- Article: not found