- Record: found
- Abstract: found
- Article: found
Ethanolamine Controls Expression of Genes Encoding Components Involved in Interkingdom Signaling and Virulence in Enterohemorrhagic Escherichia coli O157:H7
Read this article at
ABSTRACT
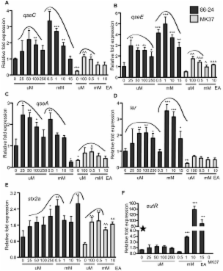
Bacterial pathogens must be able to both recognize suitable niches within the host for colonization and successfully compete with commensal flora for nutrients in order to establish infection. Ethanolamine (EA) is a major component of mammalian and bacterial membranes and is used by pathogens as a carbon and/or nitrogen source in the gastrointestinal tract. The deadly human pathogen enterohemorrhagic Escherichia coli O157:H7 (EHEC) uses EA in the intestine as a nitrogen source as a competitive advantage for colonization over the microbial flora. Here we show that EA is not only important for nitrogen metabolism but that it is also used as a signaling molecule in cell-to-cell signaling to activate virulence gene expression in EHEC. EA in concentrations that cannot promote growth as a nitrogen source can activate expression of EHEC’s repertoire of virulence genes. The EutR transcription factor, known to be the receptor of EA, is only partially responsible for this regulation, suggesting that yet another EA receptor exists. This important link of EA with metabolism, cell-to-cell signaling, and pathogenesis, highlights the fact that a fundamental means of communication within microbial communities relies on energy production and processing of metabolites. Here we show for the first time that bacterial pathogens not only exploit EA as a metabolite but also coopt EA as a signaling molecule to recognize the gastrointestinal environment and promote virulence expression.
IMPORTANCE
In order to successfully cause disease, a pathogen must be able to sense a host environment and modulate expression of its virulence genes as well as compete with the indigenous microbiota for nutrients. Ethanolamine (EA) is present in the large intestine due to the turnover of intestinal cells. Here, we show that the human pathogen Escherichia coli O157:H7, which causes bloody diarrhea and hemolytic-uremic syndrome, regulates virulence gene expression through EA metabolism and by responding to EA as a signal. These findings provide the first information directly linking EA with bacterial pathogenesis.
Related collections
Most cited references39
- Record: found
- Abstract: found
- Article: not found
A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens.
- Record: found
- Abstract: found
- Article: not found
The QseC sensor kinase: a bacterial adrenergic receptor.
- Record: found
- Abstract: found
- Article: not found