- Record: found
- Abstract: found
- Article: not found
Extensive DRB region diversity in cynomolgus macaques: recombination as a driving force
Read this article at
Abstract
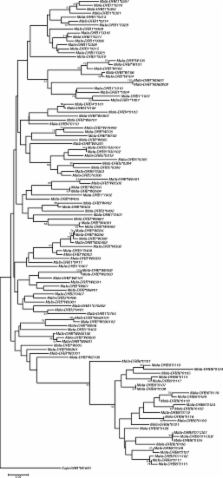
The DR region of primate species is generally complex and displays diversity concerning the number and combination of distinct types of DRB genes present per region configuration. A highly variable short tandem repeat (STR) present in intron 2 of nearly all primate DRB genes can be utilized as a quick and accurate high through-put typing procedure. This approach resulted previously in the description of unique and haplotype-specific DRB-STR length patterns in humans, chimpanzees, and rhesus macaques. For the present study, a cohort of 230 cynomolgus monkeys, including self-sustaining breeding groups, has been examined. MtDNA analysis showed that most animals originated from the Indonesian islands, but some are derived from the mainland, south and north of the Isthmus of Kra. Haplotyping and subsequent sequencing resulted in the detection of 118 alleles, including 28 unreported ones. A total of 49 Mafa-DRB region configurations were detected, of which 28 have not yet been described. Humans and chimpanzees possess a low number of different DRB region configurations in concert with a high degree of allelic variation. In contrast, however, allelic heterogeneity within a given Mafa- DRB configuration is even less frequently observed than in rhesus macaques. Several of these region configurations appear to have been generated by recombination-like events, most probably propagated by a retroviral element mapping within DRB6 pseudogenes, which are present on the majority of haplotypes. This undocumented high level of DRB region configuration-associated diversity most likely represents a species-specific strategy to cope with various pathogens.
Related collections
Most cited references49
- Record: found
- Abstract: found
- Article: not found
Mobile elements: drivers of genome evolution.
- Record: found
- Abstract: found
- Article: not found
IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex.
- Record: found
- Abstract: found
- Article: not found
