- Record: found
- Abstract: found
- Article: found
Early Cytokine Induction Upon Pseudomonas aeruginosa Infection in Murine Precision Cut Lung Slices Depends on Sensing of Bacterial Viability
Read this article at
Abstract
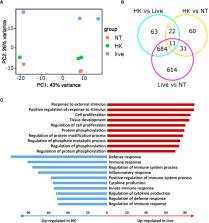
Breathing allows a multitude of airborne microbes and microbial compounds to access the lung. Constant exposure of the pulmonary microenvironment to immunogenic particles illustrates the need for proper control mechanisms ensuring the differentiation between threatening and harmless encounters. Discrimination between live and dead bacteria has been suggested to be such a mechanism. In this study, we performed infection studies of murine precision cut lung slices (PCLS) with live or heat-killed P. aeruginosa, in order to investigate the role of viability for induction of an innate immune response. We demonstrate that PCLS induce a robust transcriptomic rewiring upon infection with live but not heat-killed P. aeruginosa. Using mutants of the P. aeruginosa clinical isolate CHA, we show that the viability status of P. aeruginosa is assessed in PCLS by TLR5-independent sensing of flagellin and recognition of the type three secretion system. We further demonstrate that enhanced cytokine expression towards live P. aeruginosa is mediated by uptake of viable but not heat-killed bacteria. Finally, by using a combined approach of receptor blockage and genetically modified PCLS we report a redundant involvement of MARCO and CD200R1 in the uptake of live P. aeruginosa in PCLS. Altogether, our results show that PCLS adapt the extent of cytokine expression to the viability status of P. aeruginosa by specifically internalizing live bacteria.
Related collections
Most cited references67

- Record: found
- Abstract: found
- Article: found
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2
- Record: found
- Abstract: found
- Article: not found
Fiji: an open-source platform for biological-image analysis.

- Record: found
- Abstract: found
- Article: found