- Record: found
- Abstract: found
- Article: not found
The Uncoupling Protein 1 Gene (UCP1) Is Disrupted in the Pig Lineage: A Genetic Explanation for Poor Thermoregulation in Piglets
Read this article at
Abstract
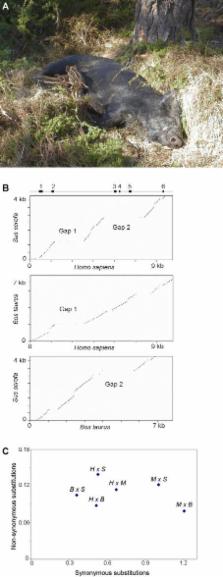
Piglets appear to lack brown adipose tissue, a specific type of fat that is essential for nonshivering thermogenesis in mammals, and they rely on shivering as the main mechanism for thermoregulation. Here we provide a genetic explanation for the poor thermoregulation in pigs as we demonstrate that the gene for uncoupling protein 1 (UCP1) was disrupted in the pig lineage. UCP1 is exclusively expressed in brown adipose tissue and plays a crucial role for thermogenesis by uncoupling oxidative phosphorylation. We used long-range PCR and genome walking to determine the complete genome sequence of pig UCP1. An alignment with human UCP1 revealed that exons 3 to 5 were eliminated by a deletion in the pig sequence. The presence of this deletion was confirmed in all tested domestic pigs, as well as in European wild boars, Bornean bearded pigs , wart hogs, and red river hogs. Three additional disrupting mutations were detected in the remaining exons. Furthermore, the rate of nonsynonymous substitutions was clearly elevated in the pig sequence compared with the corresponding sequences in humans, cattle, and mice, and we used this increased rate to estimate that UCP1 was disrupted about 20 million years ago.
Synopsis
Brown adipose tissue (BAT) is unique to mammals. It is rich in mitochondria and generates heat to maintain body temperature during cold stress, referred to as nonshivering thermogenesis. BAT is found in abundance in rodents as well as in newborns of larger mammals, including humans. Uncoupling protein 1 (UCP1) is exclusively expressed in BAT and is localized to the inner membrane of the mitochondria. Its physiological role is to uncouple oxidative phosphorylation so that most of the energy in fat stores is dissipated as heat rather than being converted to ATP.
Piglets are sensitive to cold stress and rely on shivering as the main mechanism for thermoregulation. Furthermore, pigs are the only hoofed mammals that build nests for birth; in modern pig production, heat-producing lamps are used to keep the piglets warm. It is also striking that pigs appear to lack BAT.
Here the authors show that the UCP1 gene is disrupted in domestic pigs and their wild ancestors. The inactivation of UCP1 was estimated to have happened about 20 million years ago. The finding provides an explanation for the poor thermoregulation in piglets that may have led to the evolution of the unique maternal behavior in this species.
Related collections
Most cited references16
- Record: found
- Abstract: found
- Article: not found
Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese.
- Record: found
- Abstract: found
- Article: not found
MEGA2: molecular evolutionary genetics analysis software.
- Record: found
- Abstract: found
- Article: not found
