- Record: found
- Abstract: found
- Article: found
Medulla oblongata transcriptome changes during presymptomatic natural scrapie and their association with prion-related lesions
Read this article at
Abstract
Background
The pathogenesis of natural scrapie and other prion diseases is still poorly understood. Determining the variations in the transcriptome in the early phases of the disease might clarify some of the molecular mechanisms of the prion-induced pathology and allow for the development of new biomarkers for diagnosis and therapy. This study is the first to focus on the identification of genes regulated during the preclinical phases of natural scrapie in the ovine medulla oblongata (MO) and the association of these genes with prion deposition, astrocytosis and spongiosis.
Results
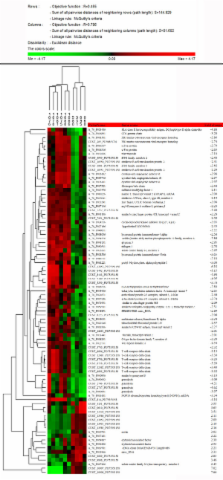
A custom microarray platform revealed that 86 significant probes had expression changes greater than 2-fold. From these probes, we identified 32 genes with known function; the highest number of regulated genes was included in the phosphoprotein-encoding group. Genes encoding extracellular marker proteins and those involved in the immune response and apoptosis were also differentially expressed. In addition, we investigated the relationship between the gene expression profiles and the appearance of the main scrapie-associated brain lesions. Quantitative Real-time PCR was used to validate the expression of some of the regulated genes, thus showing the reliability of the microarray hybridization technology.
Conclusions
Genes involved in protein and metal binding and oxidoreductase activity were associated with prion deposition. The expression of glial fibrillary acidic protein (GFAP) was associated with changes in the expression of genes encoding proteins with oxidoreductase and phosphatase activity, and the expression of spongiosis was related to genes encoding extracellular matrix components or transmembrane transporters. This is the first genome-wide expression study performed in naturally infected sheep with preclinical scrapie. As in previous studies, our findings confirm the close relationship between scrapie and other neurodegenerative diseases.
Related collections
Most cited references67
- Record: found
- Abstract: found
- Article: not found
DAVID: Database for Annotation, Visualization, and Integrated Discovery.
- Record: found
- Abstract: found
- Article: not found
PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order.
- Record: found
- Abstract: not found
- Article: not found